Summary
Herpesviruses establish life-long latent infections. During latency, gammaherpesviruses, such as Kaposi's sarcoma-associated herpesvirus (KSHV), persist as multicopy, circularized genomes in the cell nucleus and express a small subset of viral genes. KSHV latency-associated nuclear antigen (LANA) is the predominant gene expressed during latent infection. C-terminal LANA binds KSHV terminal repeat (TR) DNA to mediate DNA replication. TR DNA binding also allows tethering of the viral genome to mitotic chromosomes to mediate DNA segregation to daughter nuclei. We describe here the crystal structure of the murine gammaherpesvirus 68 LANA DNA binding domain, which is homologous to that of KSHV LANA. The structure revealed a dimer and we identified residues involved in the interaction with viral DNA. Mutation of these residues abolished DNA binding and viable latency establishment in a mouse model of infection. We also identified a positively charged patch on the dimer surface opposite to the DNA binding region and found this patch exerts an important role in the virus's ability to expand latent infection in vivo. This work elucidates the structure of the LANA DNA binding domain and identifies a novel surface feature that is critical for viral latent infection, likely by acting through a host cell protein.
Impact of LANA on Herpesvirus Latency
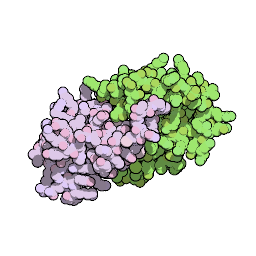
Crystal structure of the gamma-2 herpesvirus LANA DNA binding domain identifies charged surface residues which impact viral latency. PLoS Pathogens 2013 (online or pdf).
LANA structure gallery
-
 LANA-DNA model of interaction
LANA-DNA model of interaction
-

-
 LANA dimer surface
LANA dimer surface
-
 SOCS loop
SOCS loop
-
 LANA Ventral face
LANA Ventral face
-
 EBNA1 dorsal face
EBNA1 dorsal face
-
 WDR5 LANAwin interaction
WDR5 LANAwin interaction
-
 WDR5 LANA interaction
WDR5 LANA interaction
-
 Lysines residues on the Dorsal face
Lysines residues on the Dorsal face
-
 Kaposi sarcoma
Kaposi sarcoma
-
View the embedded image gallery online at:
http://mx.itqb.unl.pt/structuralvirology/index.php/blog/51-lana#sigProGalleria86d916de21
 LANA-DNA model of interaction
LANA-DNA model of interaction

 LANA dimer surface
LANA dimer surface
 SOCS loop
SOCS loop
 LANA Ventral face
LANA Ventral face
 EBNA1 dorsal face
EBNA1 dorsal face
 WDR5 LANAwin interaction
WDR5 LANAwin interaction
 WDR5 LANA interaction
WDR5 LANA interaction
 Lysines residues on the Dorsal face
Lysines residues on the Dorsal face
 Kaposi sarcoma
Kaposi sarcoma
http://mx.itqb.unl.pt/structuralvirology/index.php/blog/51-lana#sigProGalleria86d916de21



